Canine Heart Atlas:
 A data set of 8 normal canine hearts was obtained from Drs. Patrick A. Helm and Raimond L. Winslow at the Center for Cardiovascular Bioinformatics and Modeling and Dr. Elliot McVeigh at the National Institute of Health. There are two kinds of data: geometry/scalar data and diffusion tensor data. We use the template estimation method developed at the Center for Imaging Science (CIS) to generate a geometry atlas based on geometry data. Then we use LDDMM to map the diffusion tensor data from the subject to the geometry atlas. Finally, the cardiac atlas will have both geometry and diffusion tensor data, which can be used to study shape variation in a population.
A data set of 8 normal canine hearts was obtained from Drs. Patrick A. Helm and Raimond L. Winslow at the Center for Cardiovascular Bioinformatics and Modeling and Dr. Elliot McVeigh at the National Institute of Health. There are two kinds of data: geometry/scalar data and diffusion tensor data. We use the template estimation method developed at the Center for Imaging Science (CIS) to generate a geometry atlas based on geometry data. Then we use LDDMM to map the diffusion tensor data from the subject to the geometry atlas. Finally, the cardiac atlas will have both geometry and diffusion tensor data, which can be used to study shape variation in a population.
Click here to view the full directory complete with raw image data, header files, and atlases.
More information on the Analyze format used for the atlas volumes can be found under "Atlas File Formats" below.
Analysis of canine heart fiber and sheet angles:
To identify differences between fiber and sheet angles in normal and failing canine hearts, methods of computational anatomy were employed [1-3]. In this approach left ventricular (LV) cardiac fiber and sheet angles were constructed for individual ex-vivo canine heart geometry via fitting a mesh model to the epicardial and endocardial surfaces. Correspondence among different heart geometries were established by mapping individual LV geometries to a reference cardiac shape that was constructed using an iterative procedure. Each geometry first rigidly transformed to the coordinate of reference heart and then non-rigidly mapped to the reference heart using LDDMM method.
Fiber and sheet information for individual target LV geometry along with the mapping that identify corresponding locations between the target hearts and reference heart are organized as follows (Fig. 1): Click on the highlighted folders below to visit the corresponding data directories or click here to visit the exVivo/canine/CA_ANALYSIS root directory. For an explanation of the terms used in the folder hierarchy, see below fig.1.
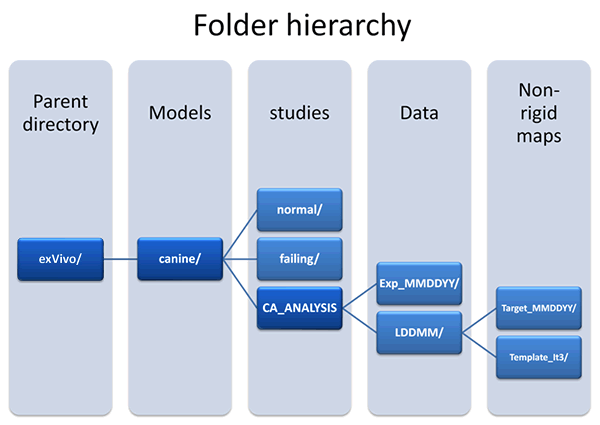
(fig 1.) folder hierarchy
- Folder Hierarchy Definitions:
- Parent directory:exVivo - Hearts removed from the animal subject.
- Models:Canine - This study utilized canine hearts.
- Studies:normal canine hearts - Adult male beagle dogs without cardiovascular disease
- Studies:failing canine hearts - Tachycardia pacing-induced model of heart failure using adult male beagle dogs
- Studies:CA_ANALYSIS - Computational anatomy analysis. This method establishes correspondence between different ventricular geometries to facilitate structural analysis between normal and disease hearts. Once all the hearts are placed in a common coordinate system, one can isolate a region of interest and identify this region on all hearts to compare cardiac fiber/sheet angle across the hearts. This method relies on mapping one heart to another using rigid and non-rigid matching algorithms. In rigid matching hearts are translated and rotated and in non-rigid matching one heart is smoothly deformed to match another heart.
- LDDMM - Non-rigid matching is conducted using large deformation diffeomorphic metric mapping (LDDMM) algorithm. (Beg M.F., M.I. Miller, A. Trouve and L. Younes (2005). "Computing Large Deformation Metric Mappings via Geodesics Flows of Diffeomorphisms." International Journal of Computer Vision, 61(2): 139-157.)
- target_MMDDYY - A ventricular canine geometry (normal or failing) that is mapped to the reference geometry (template) for regional analysis of fiber/sheet angle differences of normal and failing hearts.
- Template_it3 - A reference heart geometry that was constructed iteratively (3 iterations) using all target hearts.
- Exp_MMDDYY folder contains the following files:
- InvLDDMMSummary_MMDD.txt: Transformation for taking a point in the rigidly matched coordinate system (just prior to LDDMM) back to image space stored as test file.
- LDDMMSummary_MMDD.txt: Transformation for taking a target point to rigidly matched coordinate system (just prior to LDDMM) stored as text file.
- Rotation.txt: Rotation matrix for rigidly matching to atlas stored as text file.
- Translation.txt: Translation for rigidly matching to atlas stored as text file.
- NIH_MMDDYY_fgeo.mat: Fiber angles defined from finite element mesh (FEM) model stored as Matlab file (this file can be read to the Matlab work space using "load NIH_MMDDYY_fgeo.mat").
- NIH_MMDDYY_sgeo.mat : Sheet angles defined from finite element mesh (FEM) model stored as Matlab file.
- LDDMM folder contains two subfolders:
- Target_MMDDYY:
- Hxmap.dat, Hymap.dat, Hzmap.dat: stores non-rigid transformations (along x,y, and z directions) that map target image to the template (reference) image.
- Kixmap.dat, Kiymap.dat, Kizmap.dat: stores transformations that map template image to the target image.
- To read these file into Matlab workspace, a sample function that is called "load_dat_file.m" has been provided. To call this function, one can run:
- Kx = load_dat_file('Kixmap.dat','b'); where Kx is 3D arrays representing transformation along x component from the template geometry to the target. String 'b' indicates the byte order in which the data has been stored (b for big endian and l for little endian).
- Template_it3:
- Atlas.dat: template file constructed from both normal and failing canine hearts. This file can be read into Matlab using "load_dat_file.m" function.
- sprehh_It3.img/hdr: Atlas image stored as Analayze 7.5 format.
- To trace a region of interest that is labeled on the atlas space back to the individual heart images, one can call Matlab function named "LDDMM_FindCorrespondingFibersV1p3.m". More details are provided by executing "help LDDMM_FindCorrespondingFibersV1p3" inside Matlab. Note this function only works for identifying correspondence between Atlas.dat regions and NIH_MMDDYY.fgeo.mat and NIH_MMDDYY.sgeo.mat files. This function requires load_dat_file.m file.
- Target_MMDDYY:
- References:
- Helm PA, Younes L, Beg MF, Ennis DB, Leclercq C, Faris OP, McVeigh E, Kass D, Miller MI, Winslow RL. Evidence of structural remodeling in the dyssynchronous failing heart. Circ Res. 2006 Jan 6;98(1):125-32. Epub 2005 Dec 8. PubMed PMID:16339482.
- Helm PA, Tseng HJ, Younes L, McVeigh ER, Winslow RL. Ex vivo 3D diffusion tensor imaging and quantification of cardiac laminar structure. Magn Reson Med. 2005 Oct;54(4):850-9. PubMed PMID: 16149057; PubMed Central PMCID: PMC2396270.
- Helm P, Beg MF, Miller MI, Winslow RL. Measuring and mapping cardiac fiber and laminar architecture using diffusion tensor MR imaging. Ann N Y Acad Sci. 2005 Jun;1047:296-307. Review. PubMed PMID: 16093505.
Atlas File Formats
Atlas volumes are provided in Analyze 7.5 format. Analyze is an image processing program, written by The Biomedical Imaging resource at the Mayo Foundation. The Analyze data format is used by a wide variety of software including lddmm-volume. The specification for Analyze 7.5 is available at Analyze 7.5 file format
