Ischemic (ICM) and non-ischemic cardiomyopathy (NICM) Data
This data set contains reference shapes (templates) that are constructed at end-diastole (ED) and end-systole (ES) using high resolution (1 mm isotropic) multi detector computed tomography cardiac data from patients who were diagnosed with either ischemic (n = 13) or non-ischemic (n = 12) cardiomyopathy. The templates represent left ventricular geometry at long vertical axis view which spans from the lateral wall to the septum along the z axis (Fig. 1).
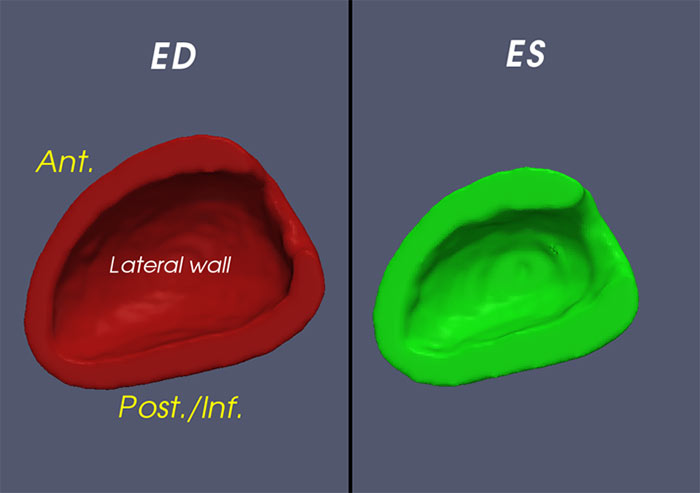
(fig 1.) Orientation of template hearts at ED and ES.
"Ant." refers to the Anterior wall of the left ventricle while "Post./Inf." refers to the posterior/inferior wall of the left ventricle.
Additional information regarding the image acquisition and post-processing methods can be found in reference [a]. Data organization is illustrated in Fig. 2. Click on the highlighted folders below to visit the corresponding data directories or click here to visit the inVivo Data root directory. For an explanation of the terms used in the folder hierarchy, see below fig.2.
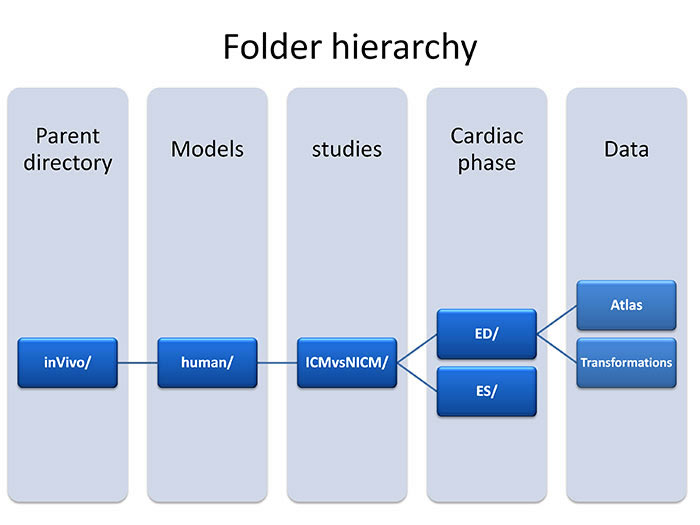
(fig 2.) data hierarchy
- Folder Hierarchy Definitions:
- Parent directory:inVivo - Study that was conducted on live human subjects.
- Models:Human - This study utilized a human heart.
- Studies:ICMvsNICM - Study compared subjects who were diagnosed with ischemic cardiomyopathy versus subjects who were diagnosed with non-ischemic cardiomyopathy.
- Cardiac phase:ED - Hearts at end-diastole cardiac phase (maximum dilation).
- Cardiac phase:ES - Hearts at end-systole cardiac phase (maximum contraction).
- Atlas : contains reference shapes that are constructed at ED and ES and stored as Analyze 7.5 format. The image resolution is 1mm x 1mm x 1mm. The ventricular geometry starts from the lateral well and terminates at the septum (z axis span).
- Transformations folder contains subfolders that are named as ICMxxx or NICMxxx, where ICMxxx represents ischemic cardiomyopathy and NICMxxx represents non-ischemic cardiomyopathy. Inside each subfolder, two files are stored that are in vtk format (http://www.vtk.org/Wiki/VTK).
- Hmap000.vtk: stores transformations that map target image to the template (reference) image.
- Kimap000.vtk: stores transformations that map template image to the target image.
- To read these file into Matlab workspace, a sample function called "LoadVtk1a.m" has been provided. To call this function, one can run:
- [Hx,Hy,Hz] = LoadVtk1a('fileName', 3, 'b'); where Hx, Hy, and Hz are 3D arrays representing transformation along x, y, and z components from the target geometry to the template, respectively. Number 3 indicates that the data is a vector quantity as opposed to scalar quantity that is represented by number 1. and string 'b' indicates the byte order in which the data has been stored (b for big endian and l for little endian).
- To transform the intensity template image into the coordinate of individual target images, one can run another Matlab function as follows: DefTemplate = LDDMMvolumeTemplate2Taget(template,Hmap); where template is a string representing the template file name (Analyze file) and Hmap is another string representing the transformation file (vtk file). DefTemplate is the deformed template image array. Note that this setup only works for the images with isotropic resolution of 1 mm.
- Also included with the downloads on this page is "LDDMMInterp3Atl2Pt.m". This is a function which interpolates the images using the LDDMM generated deformation fields that are mapping the atlas into the coordinates of patient. The deformation fields are already extracted through the "LoadVtk1a.m" matlab function. The input image (Atl) should be ordered correctly along the x-y axis (rows and columns) otherwise use perm to flip the rows and columns.
- References:
- Ardekani S, Weiss RG, Lardo AC, George RT, Lima JA, Wu KC, Miller MI, Winslow RL, Younes L. Computational method for identifying and quantifying shape features of human left ventricular remodeling. Ann Biomed Eng. 2009 Jun;37(6):1043-54. Epub 2009 Mar 26. PubMed PMID: 19322659; PubMed Central PMCID: PMC2819012.
