ExVivo Diffusion Weighted Images
Diffusion weighted images (DWI) have been collected using ex-vivo samples of canine and human hearts. Canine models contain both normal (n = 7) and failing (n = 5) hearts. Human DWI set only contains one normal heart. Each study has been named according to the following format (DTIMMDDYY) where DTI refers to diffusion tensor imaging, MM refers to month, DD refers to day , and YY refers to the year. Therefore a study that is named DTI042604 represents DTI data set that have been collected on April 26th, 2004 (Fig. 1). Click on any folder in the image below to visit the corresponding data directory or click here to visit the exVivo Data root directory.
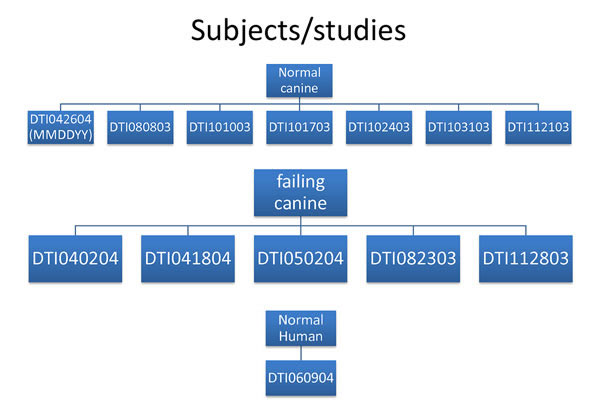
(fig 1.) dti dataset key
Additional information regarding image acquisition and post-processing methods can be found in references a-c below. Each data set directory above consists of both raw image data and processed data organised as follows (Fig. 2): For an explanation of the terms used in the folder hierarchy, see below fig.2.
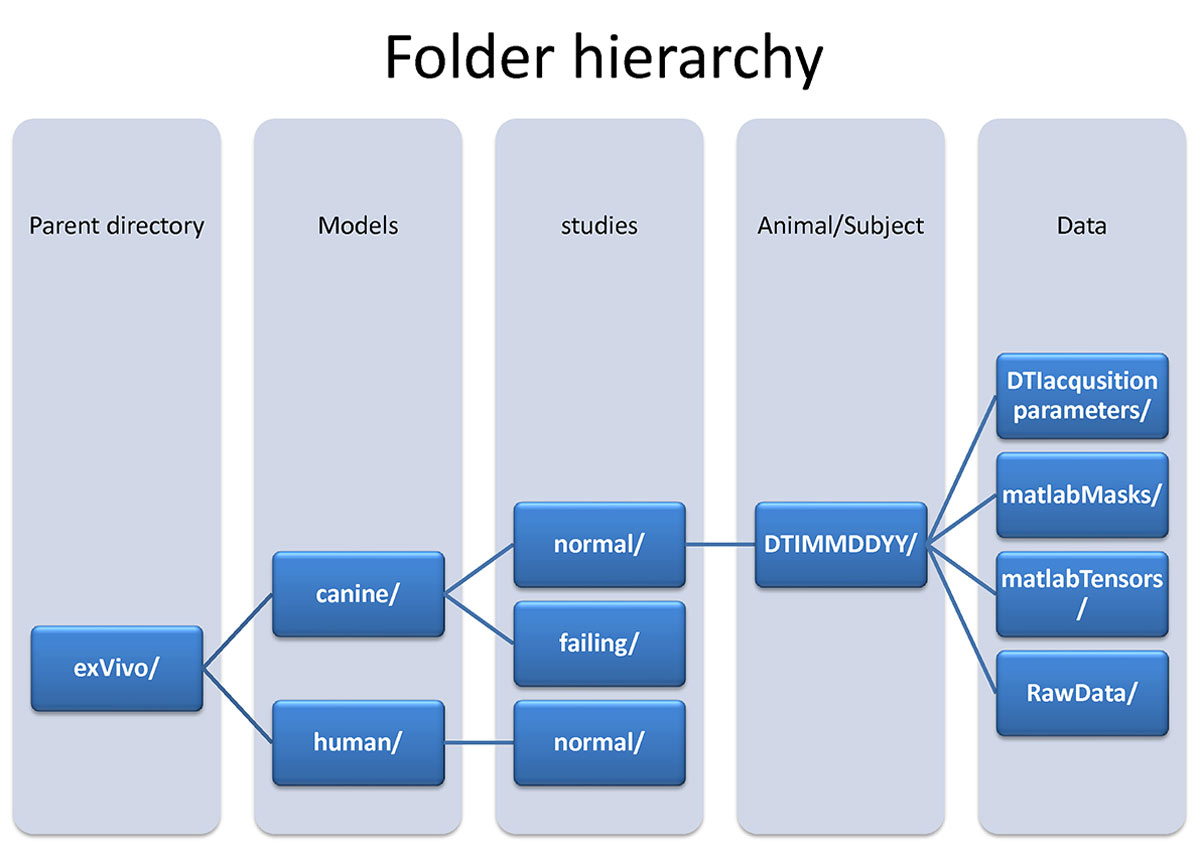
(fig 2.) data hierarchy
- Folder Hierarchy Definitions:
- Parent directory:exVivo - Hearts removed from the animal subject.
- Models:Canine - This study utilized canine hearts.
- Models:Human - This study utilized a human heart.
- Studies:normal canine hearts - Adult male beagle dogs without cardiovascular disease.
- Studies:failing canine hearts - Tachycardia pacing-induced model of heart failure using adult male beagle dogs.
- Studies:normal human heart - Adult (gender and age is missing) without cardiovascular disease.
- DTI acquisition parameters:
- contains either a text or matlab file that describes the gradient encoding directions to be used for generating diffusion tensors.
- Matlab Masks:
- contain all matlab files storing masked heart images and endo/epicardial contours. Details are:
- NIH_*_boundaries_epi.mat : A Matlab file that contains epicardium contours for the left and right ventricle in scanner coordinates.
- NIH_*_boundaries_lv.mat: A Matlab file that contains lv endocardium contours for the left ventricle in scanner coordinates.
- NIH_*_boundaries_rvfree.mat: A Matlab file that contains rv free wall endocardium contours for the left ventricle in scanner coordinates.
- NIH_*_boundaries_rvseptum.mat: A Matlab file that contains rv septal wall endocardium contours for the left ventricle in scanner coordinates.
- NIH_*_LVendo_contours.mat : A Matlab file that contains endocardium contours for the left ventricle in scanner coordinates.
- NIH_*_LVepi_contours.mat: A Matlab file that contains epicardium contours for the left ventricle in scanner coordinates.
- NIH_*_image_coordinates.mat : A Matlab file that contains image position and orientation for each slice in the scanner coordinate system.
- NIH_*_geo.mat : A Matlab 3D array whose values are a binary mask of the myocardium.
- NIH_*_painted_geo.mat: A file that contains a Matlab 3D array whose values are a coded geometry (1-LV myocardium) (2- LV) (5- RV myocardium).
- NIH_*_radgeo.mat: A file that contains a Matlab 3D array whose values are ventricular radius measurements as assessed from segmented geometry.
- NIH_*_h.mat : A file that contains a Matlab 3D array whose values are wall thickness measurements as assessed from segmented geometry.
- NIH_*_fgeo.mat: A file that contains a Matlab 3D array whose values are fiber angle measurements as assessed via the primary eigenvector.
- NIH_*_sgeo.mat : A file that contains a Matlab 3D array whose values are sheet angle measurements as assessed via the tertiary eigenvector.
- contain all matlab files storing masked heart images and endo/epicardial contours. Details are:
- Matlab Tensors:
- contain matlab files storing principal eigenvalues (e1.mat, e2.mat, e3.mat) and eigen vectors (v11.mat, x component of the primary eigenvector, …) in Euclidean coordinate system.
- Raw data:
- compressed: folder containing raw image subfolders that are acquired using diffusion encoding gradients. Each subfolder contains GE Signa.X MRI proprietary files (I.001.I.002, …) and SERHDR.00x. All the zipped files are stored in a subfolder called “zip”. To extract some basic header information, one can run a matlab function called “get_GE_Signa3xInfo.m”:
- headerInfo = get_GE_Signa3xInfo(fileName); following information has been stored in the “headerInfo”:
- offset: byte offset before start of image
- nx: Image array size in x-coordinate dimension.
- ny: Image array size in y-coordinate dimension.
- bitPerPixel: number of bits per pixel (Ex. 16).
- dz: pixel resolution along z axis.
- dx: pixel resolution along x axis.
- dy: pixel resolution along y axis.
- TLHC: 1 x 3 array representing grid coordinates for the top left hand corner.
- TRHC: 1 x 3 array representing grid coordinates for the top right hand corner.
- BRHC: 1 x 3 array representing grid coordinates for the bottom right hand corner.
- zOff: z offset from the grid origin.
- xOff: x offset from the grid origin.
- yOff: y offset from the grid origin.
- Orientation: Direction of positive x and y axis for example 'RLAP' indicates that positive x axis aligned along right to left and positive.
- TR: MR relaxation time in second, Ex: '0.5 s'.
- TE: MR echo time in ms, EX: '11.872 ms'.
- seriesNumber: Self explanatory.
- ImageNumber: Self explanatory.
- ImageDate: Self explanatory.
- ImageTime: Self explanatory.
- NIFTIdwi: contains series of 3D image volumes stored as nifti format corresponding to the diffusion weighted images in the compressed folder (Ex. DTIMMDDYY_xxx.nii). NIFTI files store in their header information regarding the correct orientation of anatomy in the scanner.
- headerInfo = get_GE_Signa3xInfo(fileName); following information has been stored in the “headerInfo”:
- compressed: folder containing raw image subfolders that are acquired using diffusion encoding gradients. Each subfolder contains GE Signa.X MRI proprietary files (I.001.I.002, …) and SERHDR.00x. All the zipped files are stored in a subfolder called “zip”. To extract some basic header information, one can run a matlab function called “get_GE_Signa3xInfo.m”:
- References:
- Helm PA, Younes L, Beg MF, Ennis DB, Leclercq C, Faris OP, McVeigh E, Kass D, Miller MI, Winslow RL. Evidence of structural remodeling in the dyssynchronous failing heart. Circ Res. 2006 Jan 6;98(1):125-32. Epub 2005 Dec 8. PubMed PMID: 16339482.
- Helm PA, Tseng HJ, Younes L, McVeigh ER, Winslow RL. Ex vivo 3D diffusion tensor imaging and quantification of cardiac laminar structure. Magn Reson Med. 2005 Oct;54(4):850-9. PubMed PMID: 16149057; PubMed Central PMCID: PMC2396270.
- Helm P, Beg MF, Miller MI, Winslow RL. Measuring and mapping cardiac fiber and laminar architecture using diffusion tensor MR imaging. Ann N Y Acad Sci. 2005 Jun;1047:296-307. Review. PubMed PMID: 16093505.
Labeled ex-vivo human heart
The files available for download below are Anlayze format containing left ventricle of ex-vivo human data labeled according to 17-segment AHA (American Heart Association) recommendation. Labels are multiplied by 10 for better visibility.
- Files
- WsH_seg_update_Labledx10.hdr (header file)
- WsH_seg_update_Labledx10.img (image file)
- References
- Standardized Myocardial Segmentation and Nomenclature for Tomographic Imaging of the Heart: A Statement for Healthcare Professionals From the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. American Heart Association Writing Group on Myocardial Segmentation and Registration for Cardiac Imaging: Manuel D. Cerqueira, Neil J. Weissman, Vasken Dilsizian, Alice K. Jacobs, Sanjiv Kaul, Warren K. Laskey, Dudley J. Pennell, John A. Rumberger, Thomas Ryan, and Mario S. Verani. Circulation 2002;105:539-542, doi:10.1161/hc0402.102975
